Validation of a novel model for the early detection of hepatocellular carcinoma
Clinical Proteomics 16, Article number: 2 (2019)
Abstract
Background
The biomarkers alpha-fetoprotein (AFP) and protein induced by vitamin K absence/antagonist-II (PIVKA-II) may be useful for detecting early-stage hepatocellular carcinoma (HCC). We evaluated the performance of AFP and PIVKA-II levels, alone and in combination with clinical factors, for the early detection of HCC.
Methods
In a case–control study, serum AFP and PIVKA-II were measured using the ARCHITECT immunoassay analyzer system in a cohort of 119 patients with HCC, 215 patients with non-malignant liver disease, and 34 healthy subjects. Five predictive models for detecting HCC were developed based on age, gender, AFP, and/or PIVKA-II levels; the best model was validated in an independent cohort of 416 patients with HCC and 412 control subjects with cirrhosis.
Results
In both cohorts, AFP and PIVKA-II concentrations were higher in patients with HCC compared to healthy controls and patients with non-malignant liver disease. The model that combined AFP and PIVKA-II, age, and gender had the highest AUC of 0.95 (0.95, 95% CI 0.93–0.98), with a sensitivity of 93% and a specificity of 84% in the development cohort, and an AUC of 0.87 (95% CI 0.85–0.90), sensitivity of 74%, and specificity of 85% in the validation cohort. When limiting the validation cohort to only early-stage HCC, the AUC was 0.85 (95% CI 0.81–0.88), sensitivity was 70%, and specificity was 86%.
Conclusions
Compared to each biomarker alone, the combination of AFP and PIVKA-II with age and gender improved the accuracy of detecting HCC and differentiating HCC from non-malignant liver disease.
Background
Worldwide, liver cancer is the fifth most common cancer in men and ninth most common in women, and the majority of primary liver cancers are hepatocellular carcinoma (HCC) [1]. The incidence of HCC has tripled between 1975 and 2011 in the US, with nearly 40,000 new cases diagnosed in 2016, primarily due to a rise in hepatitis C virus (HCV)-induced cirrhosis [1, 2]. Liver cancer is also one of the most fatal cancers, with a 5-year survival rate of 17% in the US [2] and less than 20% globally [1]. The poor prognosis of HCC is in large part related to late-stage diagnosis, as symptoms do not appear until advanced stages when there are fewer effective treatment options.
The 5-year survival rate is approximately 3% in patients with metastatic HCC [3], compared to 31% in patients with localized disease [2]. Thus, a number of clinical practice guidelines [4,5,6,7,8] recommend screening of high-risk patients, such as those with cirrhosis, to detect early-stage tumors and initiate treatment to improve outcomes [9]. Surveillance primarily involves imaging, most commonly by ultrasound with or without alpha-fetoprotein (AFP) every 6 months, as recommended by the recent guidelines from the American Association for the Study of Liver Diseases (AASLD) [2, 4]. However, early diagnosis of HCC by ultrasound alone is complicated by underlying cirrhosis and may increase the potential harms of surveillance, with low sensitivity and a high false negative rate (60%) [10]. Further, ultrasound is operator-dependent and has relatively poor reproducibility.
Circulating biomarkers may provide additional diagnostic information to complement ultrasound findings and may be particularly helpful in detecting biochemical changes associated with malignancy in the liver prior to the formation of hepatic nodules [11]. AFP is a widely used, yet imperfect biomarker for detection of liver cancer [12]. AASLD guidelines suggest that adding AFP to ultrasound may improve detection of HCC in at-risk patients with cirrhosis [4].
The biomarkers protein induced by vitamin K absence/antagonist-II (PIVKA-II), also known as des-gamma carboxyprothrombin (DCP), and AFP-L3, a glycosylated form of AFP that is more specific to liver cancer, have been investigated as additional HCC biomarkers. Abnormal carboxylation of the anticoagulation factor prothrombin by vitamin K-dependent carboxylase occurs in malignant hepatocytes, leading to increased levels of circulating PIVKA-II in patients with HCC [11, 13]. Several studies have shown that PIVKA-II has a higher sensitivity and specificity than AFP for detecting HCC versus non-malignant liver diseases [14,15,16]. However, a large, multicenter National Cancer Institute (NCI) Early Detection Research Network (EDRN) study in 836 patients reported similar areas under the receiver operating characteristic curve (ROC AUC) of 0.83 (95% CI 0.80–0.85) for AFP and 0.81 (95% CI 0.78–0.84) for PIVKA-II for differentiating between HCC and cirrhosis [17]. The same study demonstrated that the combination of AFP and PIVKA-II increased the AUC, particularly for the detection of early-stage disease [17]. PIVKA-II has been used clinically as a biomarker for risk stratification of HCC, and is now included in biomarker panels for HCC surveillance in Japanese guidelines [6, 7].
In a preliminary study of AFP and novel biomarkers for HCC using AFP and PIVKA-II assays on the Abbott ARCHITECT i2000 system, we showed that PIVKA-II had the highest diagnostic accuracy for HCC [18]. In the current study, we further evaluated the performance of the ARCHITECT AFP and PIVKA-II assays, alone and in combination with clinical factors, for the detection of HCC in populations of patients with HCC in the US, including those with early-stage HCC and non-malignant liver disease. We further validated our findings in the NCI EDRN cohort [17].
Methods
Study design and serum samples
This was a retrospective case–control study measuring the biomarkers AFP and PIVKA-II in serum samples collected between 2003 and 2016 at the Johns Hopkins Medical Institutions (JHMI) in Baltimore, MD, from patients with HCC or chronic liver disease (cirrhosis and pre-cirrhotic stages) with viral or non-viral etiology, and healthy controls. Serum samples from patients with HCC were collected prior to treatment. The study was approved by the Johns Hopkins Medicine IRB. Additional serum samples obtained after consent from patients with liver cirrhosis at the University of Texas Southwestern Medical Center (UTSMC) in Dallas, TX, were analyzed at JHMI. For each serum sample, the following de-identified data was collected: age, gender, race/ethnicity, etiology of liver disease, and HCC stage based on the TNM staging system [19], if applicable. These samples were used to develop and train the HCC detection models (development cohort).
Additional samples from the NCI EDRN cohort (validation cohort) were used to validate the best model derived from the development cohort. Validation cohort samples were obtained from EDRN [17] through an agreement with NCI. The EDRN study included 836 subjects; of these, 828 were included in this analysis (416 HCC with cirrhosis and 412 controls with cirrhosis only) and the study was powered for detecting at least a 15% sensitivity difference for a new biomarker compared with the performance of AFP alone. For each serum sample, the following de-identified data was collected: age, gender, race/ethnicity, etiology of liver disease, and HCC stage based on the Barcelona Clinic Liver Cancer (BCLC) staging system [20]. A direct comparison of the cohorts based on the TNM and BCLC systems was not possible because BCLC includes clinical criteria other than size [21].
Sample storage and assays
Serum samples were stored at approximately − 80 °C prior to analysis. US-approved AFP and ex-US‒approved PIVKA-II were measured using the ARCHITECT i2000 immunoassay analyzer (Abbott Laboratories, North Chicago, IL) per the manufacturer’s instructions [22, 23]. Each two-step sandwich immunoassay utilizes paramagnetic microparticles coated with either anti-AFP [24] or anti-PIVKA-II [25] antibodies and a chemiluminescent signal for the quantitative measurement of AFP or PIVKA-II in human serum and plasma. The performance characteristics for the ARCHITECT AFP and PIVKA-II assays are described in Table 1.
Statistical analysis
Biomarker concentrations were stratified by disease category and HCC stage. The probability of each biomarker to detect HCC was determined and Random Forest (RF) classification models were used to explore the best combination of biomarkers for the detection of HCC. RF uses a resampling method to create a large collection of de-correlated trees, and then averages them. With the RF method, the bias of the full model is equivalent to the bias of a single decision tree, but the variance is much lower due to the nature of averaging a large collection of trees [26].
All of the JHMI/UTSMC sample results, comprising the development cohort, were used to train the models. The response variable for the models was the binary HCC status (any stage HCC vs. non-HCC). Multiple RF models were developed by selecting different combinations of age, gender, and the two biomarkers as the classifiers. The best model was selected based on the combination of classifiers with the highest ROC AUC. The sensitivities (SEs) and specificities (SPs) were reported at cut-off points where the sum of the sensitivity (SE) and the specificity (SP) were maximized. The confidence intervals of AUCs and SEs/SPs were calculated based on the two-sided non-parametric method developed by Delong et al. [27].
The best, final model selected from the development cohort was assessed further. To evaluate the generalizability of the best model in a different population, an independent, blinded data set from the NCI EDRN study was used to validate model performance. The validation cohort serum samples had been previously run on a PIVKA-II sandwich immunoassay (Eisai Co, Tokyo, Japan) and an AFP immunoassay on a Wako automated system (Mountain View, CA) [17]. To address bias with the Wako and Abbott immunoassay platforms, both AFP and PIVKA-II values for the validation cohort were transformed to the ARCHITECT concentration scale as follows. One hundred EDRN matched samples (50 cases and 50 controls) were randomly selected and measured using the ARCHITECT system. The linear regression coefficients (intercept a and slope b) between the AFP and PIVKA-II values in natural log scale from the EDRN study [17] and the corresponding values measured with the ARCHITECT system were obtained for the 100 samples. The transformed values were determined by applying the regression coefficients to the AFP/PIVKA-II values in natural log scale of all validation cohort samples followed by exponential transformation. The transformed AFP/PIVKA-II values were then used for subsequent analyses.
All statistical analyses were performed using R 3.1.2 (The R Foundation for Statistical Computing).
Results
Patient demographics
The development cohort consisted of serum samples from 70 patients with stage 1 or 2 HCC, 49 patients with stage 3 or 4 HCC, and samples from 215 patients with non-malignant liver disease (40 of whom had cirrhosis) and 34 healthy subjects (Table 2). The mean age for patients in the HCC, non-malignant liver disease, and healthy control groups were 61.5, 49.5, and 58.9 years, respectively, with the majority of patients being Caucasian or African American.
Demographics of the validation cohort (n = 828) are shown in Table 3. The cohort included 416 patients with HCC, the majority of whom had BCLC stage A disease, and 412 subjects with cirrhosis. Patient age varied from 26 to 82 years, with a greater proportion of men in each group. The majority of patients were Caucasian or African American, and the majority had chronic hepatitis C (HCV) infection. The development and validation cohorts had similar demographics in terms of average age, ratio of men to women, and race/ethnicity distribution, and the majority of cases in both cohorts had a viral etiology.
Biomarker concentrations
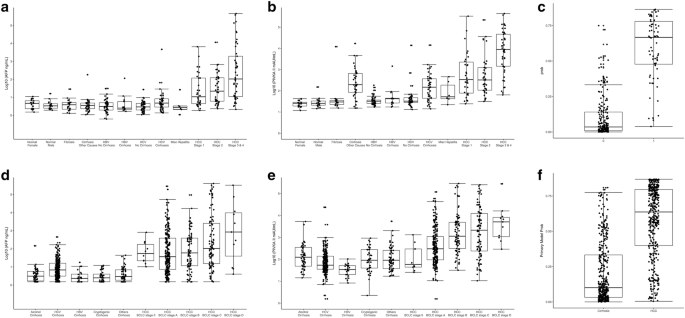
In the development cohort, AFP and PIVKA-II concentrations were found to be higher in patients with HCC than in healthy controls (p < 0.0010) and patients with chronic liver disease (p < 0.0010), with levels increasing with HCC stage (Fig. 1a, b; Additional file 1: Table S1). AFP and PIVKA-II generally demonstrated higher levels in the HCC groups than in the non-HCC and control groups, and showed increasing levels with increasing stages of HCC, as shown in a probability plot (Fig. 1c). In the validation cohort, AFP and PIVKA-II levels yielded similar patterns as seen in the development cohort (Fig. 1d, e), and the probability of the biomarker associated with the presence and staging of HCC is shown in Fig. 1f.
Concentrations of biomarkers for each study subject in the development cohort (JHMI/UTSMC) and validation cohort (NCI EDRN). a AFP concentration in the development cohort; b PIVKA-II concentration in the development cohort; c probability of HCC detection in the development cohort (no cancer = 0, cancer = 1); dAFP concentration in the validation cohort; (E) PIVKA-II concentration in the validation cohort; f probability of HCC detection in the validation cohort (cirrhosis vs. cancer)
The AFP and PIVKA-II concentrations in the validation cohort significantly correlated with the concentrations in the development cohort based on 50 control/50 HCC samples (AFP Spearman correlation coefficient ρ = 0.933, p < 0.0001; PIVKA-II Spearman correlation coefficient ρ = 0.826, p < 0.0001). However, Passing-Bablok regression and Deming regression showed systematic differences between two cohorts (data not shown). Therefore, a linear transformation method was employed to remove the systematic bias between the two cohorts.
Model performance in the development cohort
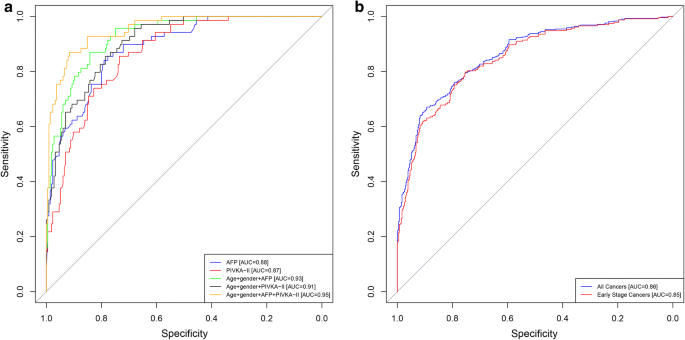
Five models were developed based on the development cohort data using age, gender, AFP, and/or PIVKA-II, and the performance of these five models were compared to each other (described in Methods). The AUC for differentiating HCC from non-malignant liver disease was similar for AFP alone (Model 1: 0.88, 95% CI 0.84–0.93) and PIVKA-II alone (Model 2: 0.87, 95% CI 0.82–0.90) (Fig. 2a). The addition of age and gender to either AFP or PIVKA-II increased the AUCs (Model 3: 0.93, 95% CI 0.90–0.96 and Model 4: 0.91, 95% CI 0.87–0.94, respectively), but the increases were not statistically significant. The best model included a combination of both biomarkers, age, and gender, which increased the AUC to 0.95 (Model 5: 95% CI 0.93–0.98), with a sensitivity of 93% and a specificity of 84%. The increase was statistically significant compared to AFP or PIVKA-II alone and either biomarker combined with age and gender (p values between 0.0000 and 0.0042). The combination of either biomarker with age and gender increased sensitivity, with only a small decrease in specificity (Table 4). When specificity was held to 90%, sensitivity reached 84%.
ROC analysis. a ROC for the development cohort (JHMI/UTSMC) for AFP (blue), PIVKA-II (red), age + gender + AFP (green), age + gender + PIVKA-II (black), age + gender + AFP + PIVKA-II (brown); b ROC for the validation cohort (EDRN) for age + gender + AFP + PIVKA-II for all cancers (blue) and for early-stage cancers (BCLC stage 0 and A; red)
Model validation in the EDRN cohort
The best model from the development cohort (model 5) was evaluated using the validation cohort data as an independent assessment of clinical performance (Table 4). Model 5, which combines both biomarkers, age, and gender, had an estimated AUC of 0.87 (95% CI 0.85–0.90) in the validation cohort. Model 5 had an estimated sensitivity of 74% and a specificity of 85%; when specificity was held to 90%, sensitivity was estimated to be 67%. When limiting the validation cohort to only early-stage HCC (BCLC stage 0 and A), the estimated AUC was 0.85 (95% CI 0.81–0.88), sensitivity was 70%, and specificity was 86% for model 5. Model 5 was further assessed in the validation cohort stratified by non-viral and viral etiologies for all cancers and early-stage cancers. AUCs were comparable for viral and non-viral cancers, though the model had a slightly lower sensitivity and higher specificity for detecting all cancers and early-stage cancers with non-viral etiology compared to those with viral etiology.
Discussion
We report here that the biomarkers AFP and PIVKA-II, when combined with age and gender, showed superior sensitivity and specificity for HCC detection compared to AFP and PIVKA-II alone or individually combined with age and gender. Further, analysis of the model in an independent validation cohort showed similar clinical performance, although with lower AUC, sensitivities, and specificities. This is important because the development cohort control group had a small number of patients with cirrhosis, while the validation cohort control group was comprised only of patients with cirrhosis. This study demonstrated the robustness of the HCC detection model with an external cohort dataset from a population of diverse composition.
Our findings are consistent with previous studies of the diagnostic accuracy of HCC biomarker panels in Asian and Western populations. In a prospective study of 734 high-risk Japanese patients with chronic hepatitis or liver cirrhosis, Ishii et al. [28] found that the combination of AFP and PIVKA-II had 65% sensitivity and 85% specificity for detecting early-stage HCC. A nested case–control study in China that included 45 patients with HCC and 138 matched controls found a similar increase in the diagnostic accuracy of the combination of AFP and PIVKA-II over either biomarker alone in patients with HCC [29]. In the US, the HALT-C trial reported that the sensitivity and specificity of PIVKA-II (74% and 46%) for the detection of early HCC were higher than those of AFP (61% and 81%), but the combination had a higher sensitivity than either biomarker alone (91%), with a 74% specificity [30]. The original EDRN case–control study of 419 US patients with HCC (208 early-stage) and 417 controls with cirrhosis found that AFP had a higher ROC AUC (0.80, 95% CI 0.77–0.84) than PIVKA-II (0.72, 95% CI 0.68–0.77), with the AUC of the combined biomarkers slightly higher than either biomarker alone (0.83, 95% CI 0.80–0.87) [17]. Our model 5 analyses in the validation cohort performed slightly better than the analyses in the original EDRN study (AUC = 0.87, 95% CI 0.85–0.90). No other biomarkers have shown better results.
One rationale for combining multiple biomarkers is that each may detect different aspects of early HCC tumor biology and provide additive information. One prospective study by Izuno et al. [31] reported that AFP was better able to detect small local tumors while PIVKA-II was more sensitive for detecting more diffuse tumors, with the combination of biomarkers having a higher diagnostic accuracy. Other studies have examined the addition of AFP-L3% as a third biomarker to improve accuracy; AFP-L3 is a glycosylated form of AFP that is specifically produced by HCC cells and has been shown to be better than AFP at differentiating between patients with HCC or cirrhosis [32]. In a study of 685 patients with HCC, 77% of patients had at least one elevated biomarker, the levels of AFP, PIVKA-II, and AFP-L3% correlated with the extent of disease as well as patient outcomes, and all three biomarkers decreased with treatment [33].
In this study, we found that PIVKA-II had lower specificity and similar sensitivity as AFP in the development cohort. Yu et al. also reported a consistently lower sensitivity of PIVKA-II compared with AFP, with similar specificities [29]; however, this is not seen consistently in the literature. Volk et al. reported that PIVKA-II is superior to either AFP or AFP-L3% at differentiating between HCC and cirrhosis (sensitivity 86%, specificity 93%), but that the AUC is lower for patients with high-risk HCC vs. low-risk HCC [34].
Differences between our findings and those of others may be related to the use of the RF model for statistical analysis, which does not specify a cut-off point, as well as differences in the patient population. When using AFP as a biomarker, a modified threshold that considers various factors, such as disease etiology/spectrum, underlying viral infection, age, and race/ethnicity for different populations may improve diagnostic accuracy [17, 35, 36]. For example, HCV is the causative agent largely responsible for the increase in incidence of HCC in the US; while HBV is the leading cause of HCC worldwide, particularly in Asia and Africa. Thus, taking viral infection into account when setting AFP biomarker thresholds may improve assessment of HCC risk in the US versus other countries.
A limitation of this study was that, given the retrospective nature of the analysis, the control groups in the development and validation cohorts were somewhat different in terms of composition and different staging systems were used, which limits assessment of specific confounders (Tables 2 and 3). A strength of this study was the demonstration of the robustness of the HCC model 5 with data from the external EDRN cohort. The large size of the validation cohort made it possible to examine the diagnostic performance of the model in subgroups of early-stage versus all cancers and viral versus non-viral etiology. The next step in the validation process is a phase 3 biomarker study using a prospective-specimen-collection, retrospective-blinded-evaluation (PRoBE) design [37], and such a study is currently underway.
Conclusions
The use of a biomarker panel of AFP and PIVKA-II in combination with age and gender improved accuracy of detecting HCC and differentiating HCC from non-malignant liver disease in a US study population as compared to the individual biomarkers alone. Additional analyses are needed to assess the diagnostic accuracy of the AFP and PIVKA-II panel for early-stage vs later-stage HCC. Further validation in a phase 3 biomarker study is needed to support the use of multiple biomarker panels to aid in the early detection of HCC.
Abbreviations
- HCC:
- hepatocellular carcinoma
- HCV:
- hepatitis C virus
- HBV:
- hepatitis B virus
- AASLD:
- American Association for the Study of Liver Diseases
- AFP:
- alpha-fetoprotein
- NCCN:
- National Comprehensive Cancer Network
- NACB:
- National Academy of Clinical Biochemistry
- PIVKA-II:
- protein induced by vitamin K absence/antagonist-II
- DCP:
- des-gamma carboxyprothrombin
- NCI:
- National Cancer Institute
- EDRN:
- Early Detection Research Network
- ROC:
- receiver operating curve
- AUC:
- area under the curve
- JHMI:
- Johns Hopkins Medical Institutions
- UTSMC:
- University of Texas Southwestern Medical Center
- BCLC:
- Barcelona Clinic Liver Cancer
- RF:
- random forest
- SE:
- sensitivity
- SP:
- specificity
References
- 1.American Cancer Society: Global cancer facts & figures, 3rd ed. Atlanta, GA: American Cancer Society. 2015; http://www.cancer.org/acs/groups/content/@research/documents/document/acspc-044738.pdf. Accessed 11 Oct 2016.
- 2.American Cancer Society: Cancer facts & figures 2016. Atlanta, GA: American Cancer Society. http://www.cancer.org/acs/groups/content/@research/documents/document/acspc-047079.pdf. Accessed 11 Oct 2016.
- 3.American Cancer Society: Liver cancer survival rates. https://www.cancer.org/cancer/liver-cancer/detection-diagnosis-staging/survival-rates.html. Accessed September 14, 2017.
- 4.Heimbach JK, Kulik LM, Finn RS, Sirlin CB, Abecassis MM, Roberts LR, et al. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology. 2018;67:358–80.
- 5.European Organisation For Research and Treatment Of Cancer. EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2012;56:908–43.
- 6.Kokudo N, Hasegawa K, Akahane M, Igaki H, Izumi N, Ichida T, et al. Evidence-based clinical practice guidelines for hepatocellular carcinoma: The Japan Society of Hepatology 2013 update (3rd JSH-HCC Guidelines). Hepatol Res. 2015;45:123–7.
- 7.Makuuchi M, Kokudo N, Arii S, Futagawa S, Kaneko S, Kawasaki S, et al. Development of evidence-based clinical guidelines for the diagnosis and treatment of hepatocellular carcinoma in Japan. Hepatol Res. 2008;38:37–51.
- 8.National Comprehensive Cancer Network: NCCN clinical practice guidelines in oncology: hepatobiliary cancers. 2016; https://www.nccn.org/professionals/physician_gls/pdf/hepatobiliary.pdf. Accessed 11 Oct 2016.
- 9.Singal AG, Pillai A, Tiro J. Early detection, curative treatment, and survival rates for hepatocellular carcinoma surveillance in patients with cirrhosis: a meta-analysis. PLoS Med. 2014;11:e1001624.
- 10.Atiq O, Tiro J, Yopp AC, Muffler A, Marrero JA, Parikh ND, et al. An assessment of benefits and harms of hepatocellular carcinoma surveillance in patients with cirrhosis. Hepatology. 2017;65:1196–205.
- 11.Tsuchiya N, Sawada Y, Endo I, Saito K, Uemura Y, Nakatsura T. Biomarkers for the early diagnosis of hepatocellular carcinoma. World J Gastroenterol. 2015;21:10573–83.
- 12.Li D, Satomura S. Biomarkers for hepatocellular carcinoma (HCC): an update. Adv Exp Med Biol. 2015;867:179–93.
- 13.Liebman HA, Furie BC, Tong MJ, Blanchard RA, Lo KJ, Lee SD, et al. Des-gamma-carboxy (abnormal) prothrombin as a serum marker of primary hepatocellular carcinoma. N Engl J Med. 1984;310:1427–31.
- 14.Marrero JA, Su GL, Wei W, Emick D, Conjeevaram HS, Fontana RJ, et al. Des-gamma carboxyprothrombin can differentiate hepatocellular carcinoma from nonmalignant chronic liver disease in american patients. Hepatology. 2003;37:1114–21.
- 15.Li C, Zhang Z, Zhang P, Liu J. Diagnostic accuracy of des-gamma-carboxy prothrombin versus alpha-fetoprotein for hepatocellular carcinoma: a systematic review. Hepatol Res. 2014;44:E11–25.
- 16.Pote N, Cauchy F, Albuquerque M, Voitot H, Belghiti J, Castera L, et al. Performance of PIVKA-II for early hepatocellular carcinoma diagnosis and prediction of microvascular invasion. J Hepatol. 2015;62:848–54.
- 17.Marrero JA, Feng Z, Wang Y, Nguyen MH, Befeler AS, Roberts LR, et al. Alpha-fetoprotein, des-gamma carboxyprothrombin, and lectin-bound alpha-fetoprotein in early hepatocellular carcinoma. Gastroenterology. 2009;137:110–8.
- 18.Morota K, Nakagawa M, Sekiya R, Hemken PM, Sokoll LJ, Elliott D, et al. A comparative evaluation of Golgi protein-73, fucosylated hemopexin, alpha-fetoprotein, and PIVKA-II in the serum of patients with chronic hepatitis, cirrhosis, and hepatocellular carcinoma. Clin Chem Lab Med. 2011;49:711–8.
- 19.American Joint Committee on Cancer. AJCC cancer staging manual. 7th ed. New York: Springer; 2010.
- 20.Llovet JM, Bru C, Bruix J. Prognosis of hepatocellular carcinoma: the BCLC staging classification. Semin Liver Dis. 1999;19:329–38.
- 21.Maida M, Orlando E, Camma C, Cabibbo G. Staging systems of hepatocellular carcinoma: a review of literature. World J Gastroenterol. 2014;20:4141–50.
- 22.ARCHITECT PIVKA-II [package insert]. Weisbaden, Germany: Abbott GmbH & Co. KG; 2015.
- 23.ARCHITECT AFP [package insert]. Sligo, Ireland: Abbott Ireland; 2013.
- 24.Morota K, Komori M, Fujinami R, Yamada K, Kuribayashi K, Watanabe N, et al. Improvement and multicenter evaluation of the analytical performance of an automated chemiluminescent immunoassay for alpha fetoprotein. Int J Biol Markers. 2012;27:39–46.
- 25.Fujita K, Kinukawa H, Ohno K, Ito Y, Saegusa H, Yoshimura T. Development and evaluation of analytical performance of a fully automated chemiluminescent immunoassay for protein induced by vitamin K absence or antagonist II. Clin Biochem. 2015;48:1330–6.
- 26.Breiman L. Random forests. Mach Learn. 2001;45:5–32.
- 27.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–45.
- 28.Ishii M, Gama H, Chida N, Ueno Y, Shinzawa H, Takagi T, et al. Simultaneous measurements of serum alpha-fetoprotein and protein induced by vitamin K absence for detecting hepatocellular carcinoma. South Tohoku District Study Group. Am J Gastroenterol. 2000;95:1036–40.
- 29.Yu R, Xiang X, Tan Z, Zhou Y, Wang H, Deng G. Efficacy of PIVKA-II in prediction and early detection of hepatocellular carcinoma: a nested case-control study in Chinese patients. Sci Rep. 2016;6:35050.
- 30.Lok AS, Sterling RK, Everhart JE, Wright EC, Hoefs JC, Di Bisceglie AM, et al. Des-gamma-carboxy prothrombin and alpha-fetoprotein as biomarkers for the early detection of hepatocellular carcinoma. Gastroenterology. 2010;138:493–502.
- 31.Izuno K, Fujiyama S, Yamasaki K, Sato M, Sato T. Early detection of hepatocellular carcinoma associated with cirrhosis by combined assay of des-gamma-carboxy prothrombin and alpha-fetoprotein: a prospective study. Hepatogastroenterology. 1995;42:387–93.
- 32.Sato Y, Nakata K, Kato Y, Shima M, Ishii N, Koji T, et al. Early recognition of hepatocellular carcinoma based on altered profiles of alpha-fetoprotein. N Engl J Med. 1993;328:1802–6.
- 33.Toyoda H, Kumada T, Kiriyama S, Sone Y, Tanikawa M, Hisanaga Y, et al. Prognostic significance of simultaneous measurement of three tumor markers in patients with hepatocellular carcinoma. Clin Gastroenterol Hepatol. 2006;4:111–7.
- 34.Volk ML, Hernandez JC, Su GL, Lok AS, Marrero JA. Risk factors for hepatocellular carcinoma may impair the performance of biomarkers: a comparison of AFP, DCP, and AFP-L3. Cancer Biomark. 2007;3:79–87.
- 35.Gopal P, Yopp AC, Waljee AK, Chiang J, Nehra M, Kandunoori P, et al. Factors that affect accuracy of alpha-fetoprotein test in detection of hepatocellular carcinoma in patients with cirrhosis. Clin Gastroenterol Hepatol. 2014;12:870–7.
- 36.Richardson P, Duan Z, Kramer J, Davila JA, Tyson GL, El-Serag HB. Determinants of serum alpha-fetoprotein levels in hepatitis C-infected patients. Clin Gastroenterol Hepatol. 2012;10:428–33.
- 37.Pepe MS, Feng Z, Janes H, Bossuyt PM, Potter JD. Pivotal evaluation of the accuracy of a biomarker used for classification or prediction: standards for study design. J Natl Cancer Inst. 2008;100:1432–8.
Authors’ contributions
PMH, LJS, and GJD conceived and designed the study, collected and analyzed data, and helped write the manuscript; XY, JD, and ML analyzed the data and helped write the manuscript; DE collected and analyzed data and revised the manuscript; SHG and JAM collected and analyzed data and helped write the manuscript; and ZF, SS, and DWC designed the study and helped write the manuscript. All authors read and approved the final manuscript.
Acknowledgements
The authors acknowledge the contributions of NCI EDRN investigators who participated in the collection of specimens, including Yinghui Wang, Mindie Nguyen, Alex Befeler, Lewis Roberts, K. Rajender Reddy, Denise Harnois, Josep Llovet, Daniel Normolle, Jackie Dalhgren, David Chia, Anna Lok, Paul Wagner, and Myron Schwartz. Stacey Tobin, PhD, received compensation for her assistance in preparing this manuscript.
Preliminary data reported in this manuscript were previously presented at the 43th ISOBM Annual Congress in Chicago, Illinois, September 2, 2016.
Competing interests
Philip M. Hemken is an employee of, owns stock in, and holds intellectual property interests with Abbott Laboratories; Lori J. Sokoll receives research funding support from Abbott Laboratories; At time of contributions to study, Xiaoqing Yang was an employee of, received research funding support from, and held intellectual property interests with Abbott Laboratories; Jianliang Dai has nothing to disclose; Debra Elliott has nothing to disclose; Susan H. Gawel is an employee of, owns stock in, receives research funding support from, and holds intellectual property interests with Abbott Laboratories; Michael Lucht is an employee of Abbott GmbH & Co. KG and owns stock in Abbott Laboratories; Ziding Feng has nothing to disclose; Jorge A. Marrero has nothing to disclose; Sudhir Srivastava has nothing to disclose; Daniel W. Chan has received research funding support from Abbott Laboratories; Gerard J. Davis is an employee of, owns stock in, and receives research funding support from Abbott Laboratories.
Availability of data and materials
The dataset from JHMI used to develop the model is available from the corresponding author upon request. The dataset from EDRN used to validate the model is not publicly available as it is a consortium reference set from an ongoing trial and blinding must be maintained.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The study was approved by the Johns Hopkins Medicine IRB.
Funding
The study was supported by Abbott Laboratories and the National Cancer Institute (NCI) Early Detection Research Network (EDRN) (NIH U24 CA115102).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Δεν υπάρχουν σχόλια:
Δημοσίευση σχολίου